BioGPS
The structural comparison of protein binding sites is increasingly important in drug design; identifying structurally similar sites can be useful for techniques such as drug repurposing, and also in a polypharmacological approach to deliberately affect multiple targets in a disease pathway, or to explain unwanted off-target effects. Once similar sites are identified, identifying local differences can aid in the design of selectivity. Such an approach moves away from the classical "one target one drug" approach and toward a wider systems biology paradigm.
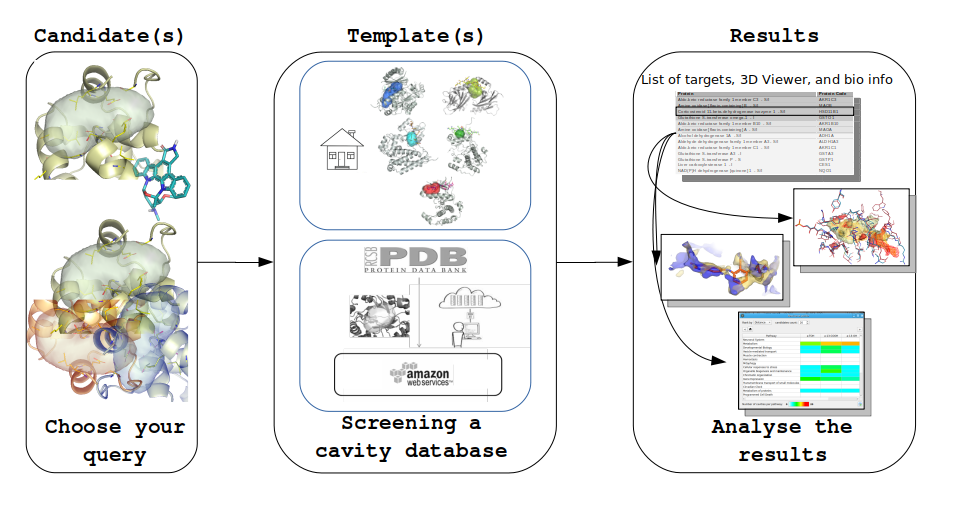
BioGPS, that is based on the software FLAP which combines GRID Molecular Interactions Fields (MIFs) and pharmacophoric fingerprints, enables pocket-pocket virtual screening. BioGPS comprises the automatic preparation of protein structure data, identification of binding sites, and subsequent comparison by aligning the sites and directly comparing the MIFs. Chemometric approaches are included to reduce the complexity of the resulting data on large datasets, enabling users to focus on the most relevant information. BioGPS comes with a curated database of ~800,000 pockets identified from publicly available structures, with their MIFs pre-computed and their biological annotation..
Key Features
- automatic protein preparation
- identification of pockets
- pocket-pocket comparison
- integrated chemometric and clustering approaches
- curated database of ~800,000 pockets
- biological annotation of pockets
- support for large computation using cloud CPUs
Key applications
- protein pocket comparison
- protein pocket clustering
- drug repurposing
- ligand exchange
- off-targeting prediction
- ligand selectivity design
Comparison and clustering including the (a) automatic preparation of protein structure data, (b) identification of pockets, (c) subsequent comparison by aligning the sites and directly comparing the MIFs, (d) PCA, PLS, Kmeans and hierarchical clustering tools.
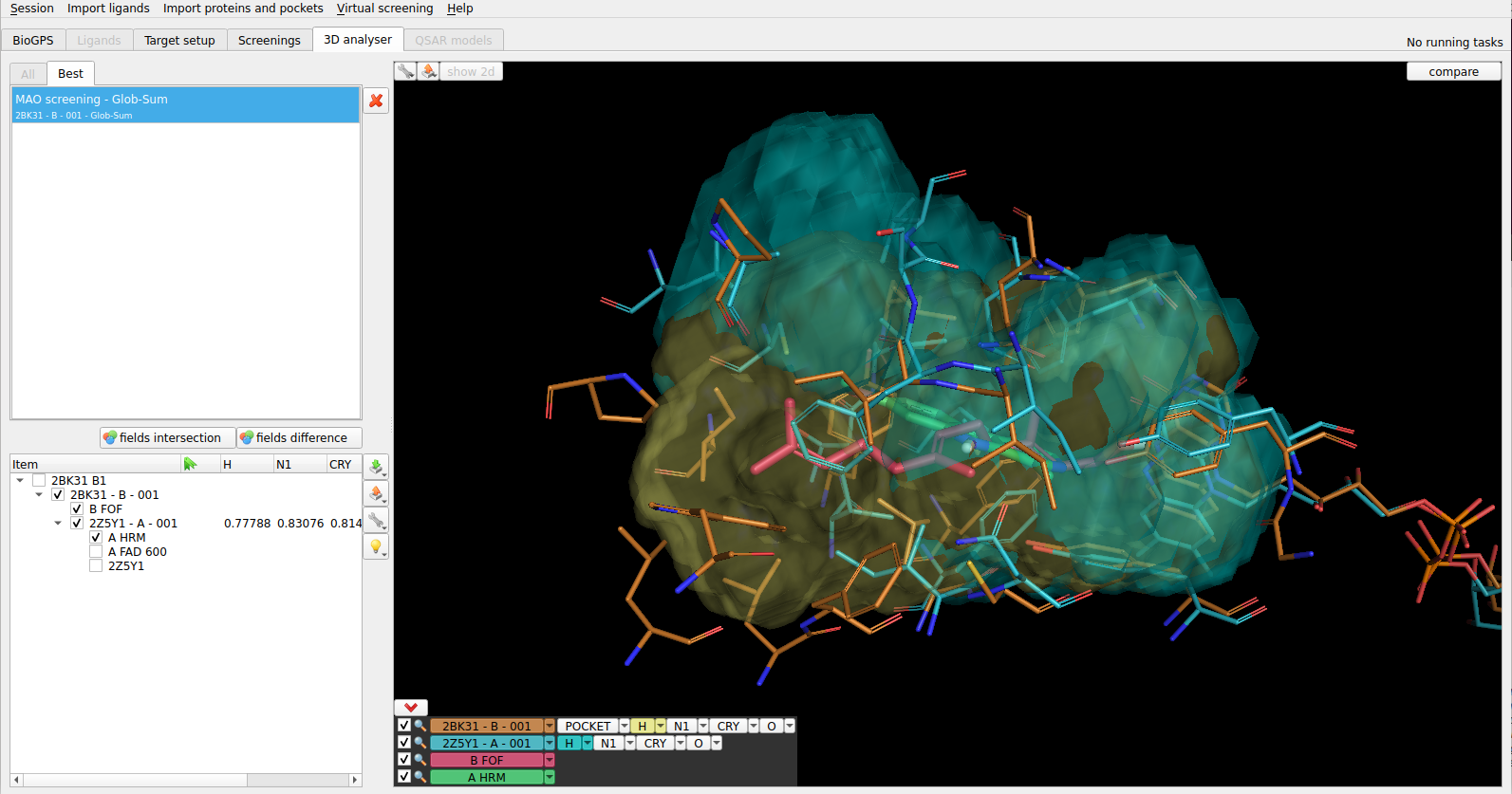
Figure 1. Superposition of two protein pockets
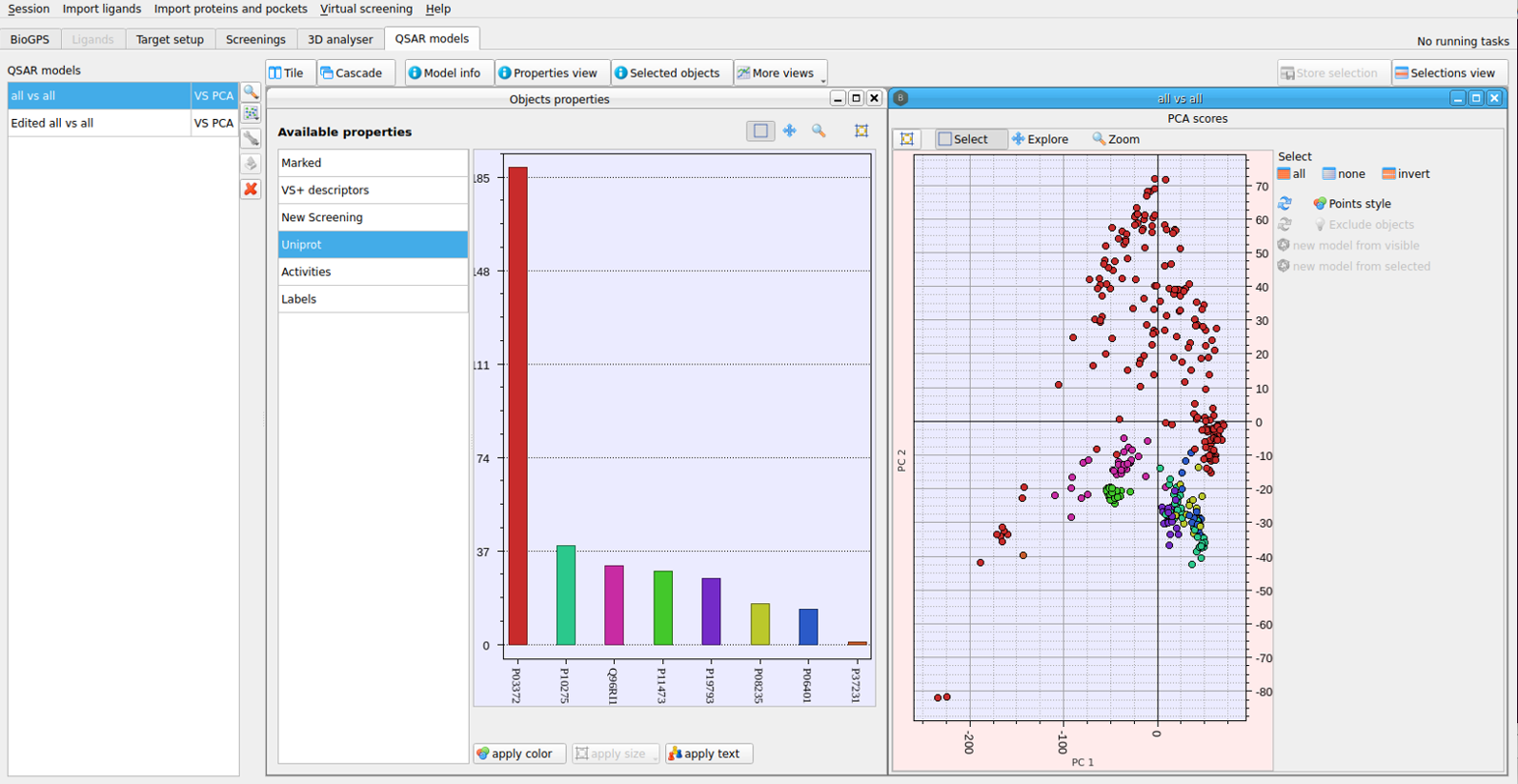
Figure 2. Principal Component Analysis (PCA) applied to a Pocketome subset
Pocketome, a curated database of pockets detected using high quality protein structures from the Protein Data Bank. It includes ~800,000 pockets ready for large scale virtual screening
- 120,506 biological units - 38,078 biological units in the non-redundant version (best X-ray resolution)
- 199,485 chains - 40,201 chains in the non-redundant version (best X-ray resolution)
- 34,885 unique proteins (Uniprot codes)
- 867,511 pockets - 304,335 pockets in the non-redundant version (best X-ray resolution)
- 111,479 liganded pockets
- 24,156 ligands (~6K peptides)
- 5,700 organisms
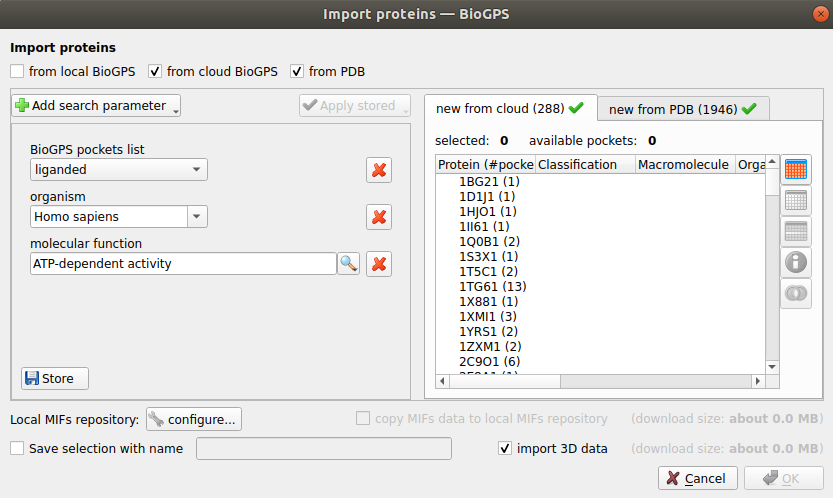
Figure 3. Example of pocket selection from Pocketome
Metadata, a set of chemical and biological annotation for each pocket in the Pocketome, helping filtering, subset creation and results rationalization.
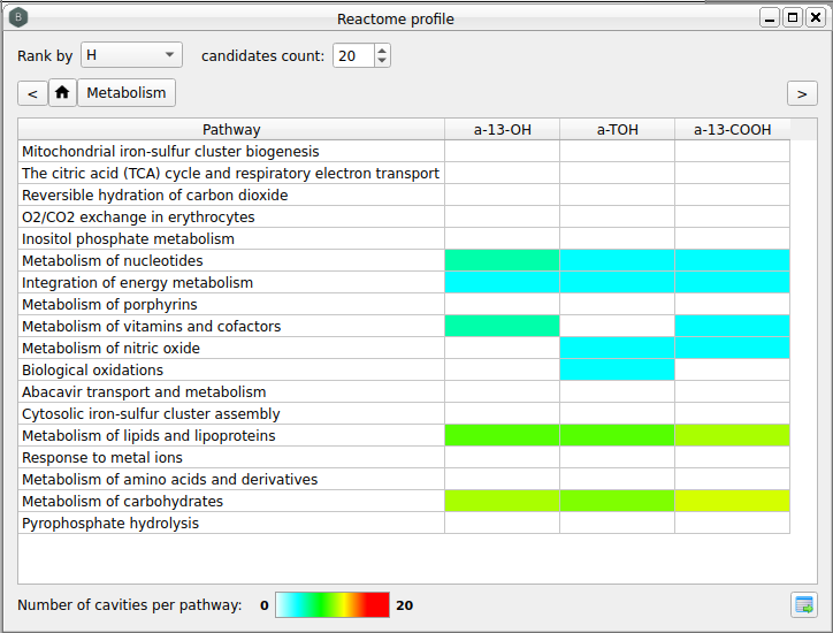
Figure 4. Reactome pathways profiling
Runaway, a web service allowing to send large computation on cloud CPUs. Both Molecular Discovery Ltd. or an internal cloud can be used.
BioGPS success stories
- BioGPS has been used for developing ELIOT (E3 LIgase pocketOme navigaTor), an accurate and complete platform containing the E3 ligase pocketome for enabling navigation to aid selection of new E3 ligases and new ligands for the design of new PROTACs.
- BioGPS has been used for building CROMATIC (CROss-relationship MAp of CaviTIes from Coronaviruses). CROMATIC encloses both a comprehensive and a non-redundant version of the cavities collection from Coronaviruses, and a similarity map revealing, on the one hand, cavities that are conserved, and, on the other hand, unexpected similarities for multi-target therapy strategies. The CROMATIC repository is freely available to scientific community at https://github.com/moldiscovery/sars-cromatic.
- BioGPS has been used for detecting and characterizing protein pockets for HotSpot's proprietary SpotFinderTM platform.
- In collaboration with Nestlè Research Center, we investigated the ligand-protein binding screening across the full proteome to identify new molecular initiating events (MIEs) for food chemicals.
- BioGPS/FLAPdock integrated drug-repurposing pipeline was successfully used for identifying new compounds active in the low-micromolar range, as thymidylate synthase inhibitors.
- GPCRome: GPCRs binding site platform for similarity assessments and screening/ligand repurposing efforts, built in collaboration with SoseiHeptares
REFERENCES
ALTEX. 2020;37(1):85-94.
