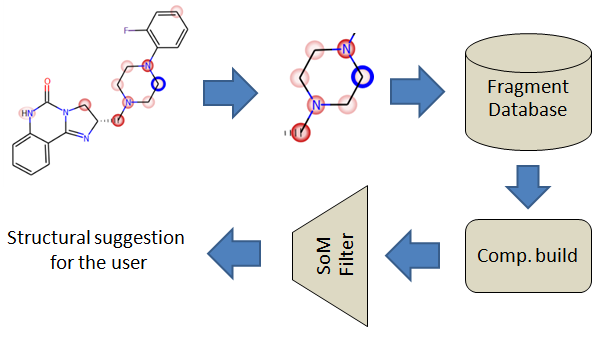
MetaDesign
MetaDesign is a tool designed to find bioisosteric replacement of fragments with the intent to produce molecules with improved metabolic properties.
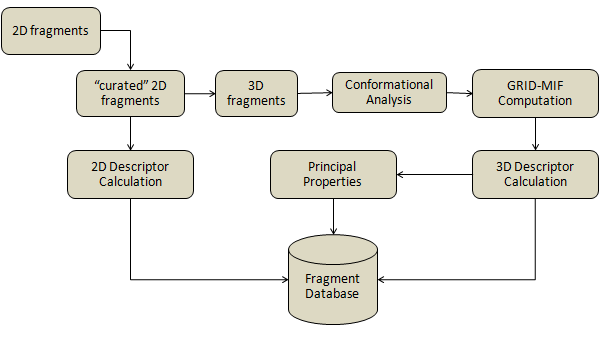
Personalized databases can be built by using the MetaDesign database builder command line tools here below:
To add fragments to the database, they have to be written in SD file format and should include 1 to 5 Dummy atoms (using symbol Du) that will be used as anchor points. The SD file can then be provided as an argument for the command-line tool. This is an example of the SD input file:
All the bioisosteric descriptors used by MetaDesign will be prepared and stored in the database with the structure of the fragments and their possible conformers.
The command-line tool will create a new database file named scaffoldDB if it's not already present in working directory, otherwise fragments will be appended to the existing database file. Once the database is built it can be freely renamed and used by the MetaDesign module of Metasite or WebMetabase.
Some pre-computed databases of building blocks from commercial catalogs are available for download here below:
