ELIOT
E3 LIgase pocketOme navigaTor
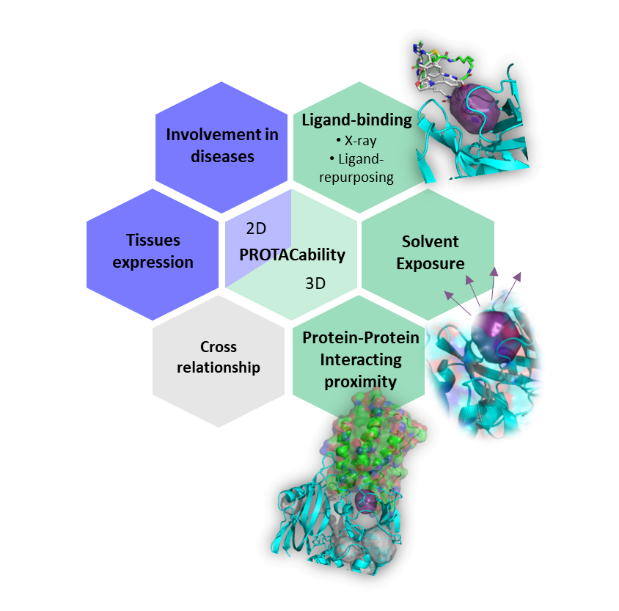
Selection of the appropriate E3 ligase for a PROTAC is clearly an important part of PROTAC design, and it may be important to vary the E3 ligase as resistance in cancer cells has been detected following chronic treatment with PROTACs based on VHL or CRBN. The biological function, level of expression in the tissues, cell localization and the degree of involvement in human diseases of the E3 ligase are all important factors to consider; recruiting E3 ligases with a selective tissue expression profile presents a unique opportunity for some therapeutic applications, especially if there is a difference between diseased tissue versus healthy tissue. The opportunity to engage new E3 ligases with a selective tissue expression profile, that can hijack and control the UPS (Ubiquitin Proteasome System), could greatly extend the applicability of degrading molecules as new alternative therapeutic modality.
ELIOT (E3 LIgase pocketOme navigaTor) (1) is a platform containing an E3 ligase pocketome enabling navigation for the selection of new E3 ligases and new ligands for the design of new PROTACs. We have collected all available human crystallographic structures and using the BioGPS(2) approach we have identified, characterized and described all possible pockets on the surface of the E3 ligases with the aim of unveiling new sites suitable for binding PROTACs. Each pocket is characterized with several descriptors and information coming from in-house tools and public resources. The result is a navigable E3 ligases pocketome equipped with various information to facilitate selection of the most appropriate E3 ligase. ELIOT contains 204 different genes for a total of 1038 structures, 2517 pockets and 1039 regions.
Key Features
- Interactive and intuitive GUI
- Gene ID card
- 3D structural information:
- PROTAC-ability score (combination of Ligand-binding score, solven exposure score and Protein- Protein interacting proximity score)
- Representativity score to define conserved, middle and transient regions
- Clustering analysis based on residues to define all possible regions
- E3 pocketome cross-relationship analysis based on GRID MIFs
- Similarity analysis based on GRID MIFS between E3 pockets and known liganded pockets from the PDB in order to provide ligand opportunities
- Meta-data: subfamilies classification, E3 ligase expression in human tissues and E3 ligase level of expression in cancers
Key applications
- Identification of new interesting binding site for PROTAC design
- Identification of new E3 ligase for PROTAC design
- Identification of new possible E3 ligand for PROTAC design
- Ligand/scaffold repurposing applications
- E3 pocket comparison
- E3 off-targeting prediction
REFERENCES
- Palomba T, Cross S, Baroni M, Cruciani G, Siragusa L. ELIOT: A platform to navigate the E3 pocketome and aid the design of new PROTACs. Chem Biol Drug Des. 2022;(May):1-18.
- Siragusa L, Cross S, Baroni M, Goracci L, Cruciani G. BioGPS: Navigating biological space to predict polypharmacology, off-targeting, and selectivity. Proteins Struct Funct Bioinforma. 2015;83(3):517-32.
