LipostarMSI
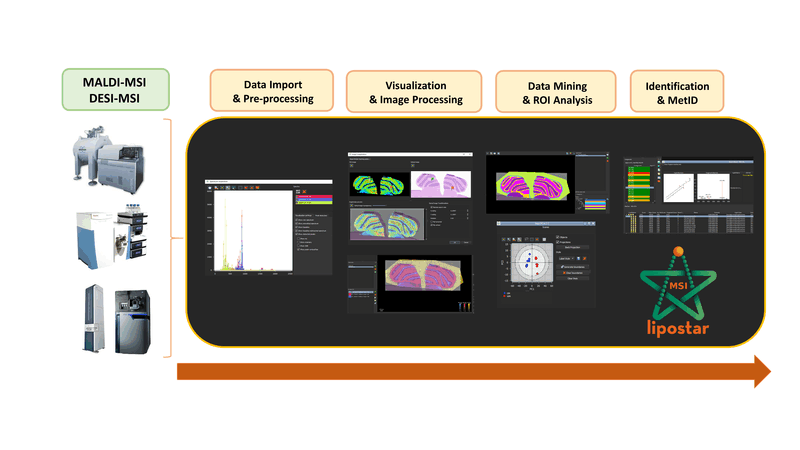
LipostarMSI is a comprehensive, vendor-neutral software for mass spectrometry imaging (MSI) that supports both untargeted and targeted data analysis.
The workflow supported by LipostarMSI covers all the steps required for MSI data analysis: raw data processing, histological co-registration, manual drawing or importing of regions annotated by pathologists, image visualization, a variety of univariate and multivariate image and spectral analysis tools, co-localization analysis, and automated lipids/metabolite annotation, including MS/MS-based identification.
A revolution in MSI for pharmaceutical and medical applications
- Both targeted and untargeted MSI data analysis supported, greatly streamlining biochemical interpretation of the data
- Customizable peak picking, pixel spectra alignment, data reduction
- Multiple images co-registration
- Automatic import of ROI annotated by expert pathologists using QuPath and Pannoramic Viewer
- Automatic metabolite and drug metabolite annotation
- Automatic MS/MS-based lipid identification
- Automatic report generation
Features
- Reading of the imzML file format
- Import of MS/MS spectra from LC-MS acquisitions. The following LC-MS formats are supported:
- Agilent Q-Tof(*.d): AutoMS and full scan at multiple energies of collision.
- Waters (*.raw): MSe and MSMS (MassLynx needs to be installed).
- Thermo-Fisher (*.RAW): Ion-Trap and Orbitrap Data Dependent Scan, Exactive, Q-Exactive.
- ABSciex *.wiff file format.
- Bruker (*.d): QTof data dependent scan.
- Peak detection, spectra calibration, alignment and filtering
- MSI File preview to fine-tune import settings
- Multi dataset support
- Instant/real-time visualization of single and multiple (overlayed) ion density images
- Pixel spectrum and ROI average spectrum visualization
- Manual ROI definition
- Import of ROI annotations from Pannoramic Viewer and QuPath
- Co-registration and transparency of multiple optical images for each dataset
- Denoise, contrast enhancement, normalization of ion density images
- Customizable multi dataset visualization layout
- Spatial segmentation
- Biomarker driven spatial segmentation
- Co-localization analysis based on ROIs, m/z values and segmentation clusters
- PCA
- Background processing of computational intensive tasks
- Easy, quick and automatic report generation
- Semi-automatic out-of-tissue (background) compounds exclusion/subtraction
- Preferred list of m/z values management
- Lipid Identification and customizable Lipid Database (support for MS/MS based identification when available)
- Automatic drug metabolites identification
- ROI Analysis (supervised and unsupervised multivariate statistical analysis)
- Algorithms optimized and parallelized to fully exploit the features of the most recent multi-core hardware architectures
- Module (the DB Manager) to build customized databases of fragmented lipids, including stable isotope-labeled species, and of any metabolite of interest
What users think
“The attention that Molecular Horizon have paid to the needs of the MSI community and their personal interactions with the user-base have led to the development of a highly versatile software in LipostarMSI – much needed in the MSI field.”
– Prof. Ruth Andrew
Chair of Pharmaceutical Endocrinology
University/BHF Centre for Cardiovascular Science
Queen’s Medical Research Institute
University of Edinburgh
“Fast, comprehensive and user-friendly software that saves you time and exasperations!”
– Klára Ščupáková, MSc
PhD student
M4I Institute, Division of Imaging Mass Spectrometry
University of Maastricht
“I am very happy I found LipostarMSI for the analysis of our untargeted imaging mass spectrometry lipidomics datasets. Molecular Horizon is determined to offer a software that contains everything scientists need to completely analyse their datasets. You can visualise multiple datasets at the same time, use unsupervised clustering algorithms based on the image data to look for differences between tissue areas, further identify m/z values of interest using analyses based on user set regions of interest and even identify lipids (or drug metabolites) all in the same software. If you need a special feature that is not part of the software yet, Molecular Horizon listens to your wishes and often implements a useful tool that fits your needs very quickly. I would highly recommend LipostarMSI for the analysis of imaging mass spectrometry datasets.”
– Luise A. Seeker
Postdoctoral Research Fellow
MRC Scottish Centre for Regenerative Medicine
Edinburgh BioQuarter
Availability
LipostarMSI is now available, for all inquires please refer to Molecular Horizon
